Docs to table: Building a Streamlit App to Extract Tables from PDFs and Answer Questions
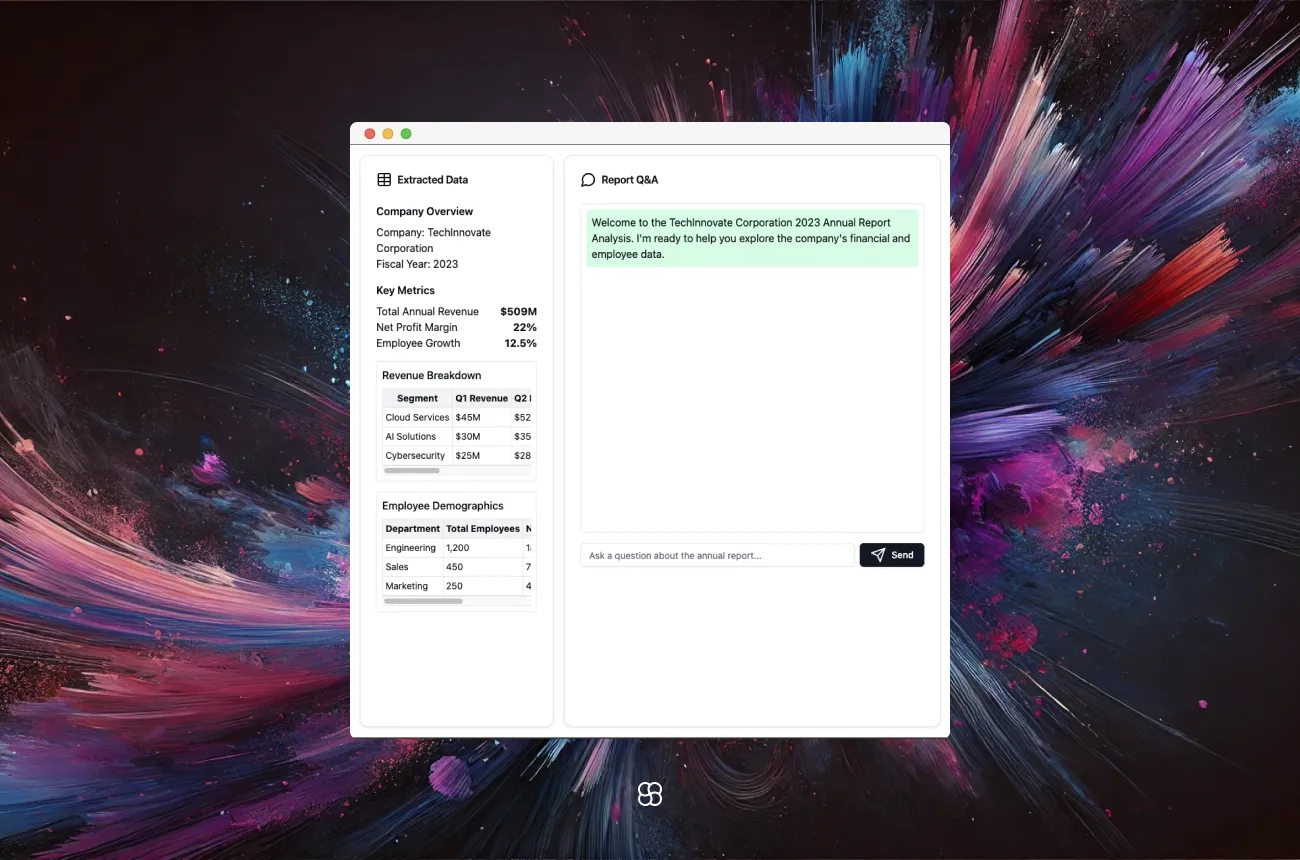
In this blog post, we'll walk through the process of building a Streamlit app that allows users to upload a PDF file, extract tables from it, and then ask questions about the content which the app will answer using natural language processing techniques. We'll be using several popular Python libraries including Streamlit, Unstructured, LangChain, and ChromaDB.
Requirements
To follow along, you'll need Python installed as well as the following libraries:
- streamlit
- unstructured
- langchain
- chromadb
- openai
- tiktoken
You can install them using pip:
pip install streamlit unstructured langchain chromadb openai tiktokenMake sure you also have an OpenAI API key as we'll be using their language model.
Extracting Tables from PDFs
The first step is to allow the user to upload a PDF file and extract any tables it contains. We'll use the Unstructured library for this.
import tempfile
from unstructured.partition.pdf import partition_pdf
from unstructured.staging.base import elements_to_json
def process_json_file(input_filename):
# Read the JSON file
with open(input_filename, 'r') as file:
data = json.load(file)
# Iterate over the JSON data and extract required table elements
extracted_elements = []
for entry in data:
if entry["type"] == "Table":
extracted_elements.append(entry["metadata"]["text_as_html"])
# Write the extracted elements to the output file
with open(f"outputs/{input_filename.split('/')[-1]}.txt", 'w') as output_file:
for element in extracted_elements:
output_file.write(element + "\\n\\n") # Adding two newlines for separation
def extract_tables_to_docs(filename, output_dir, strategy="hi_res", model_name="yolox", chunk_size=1000, chunk_overlap=200):
# Partition the PDF and extract table structure
elements = partition_pdf(filename=filename, strategy=strategy, infer_table_structure=True, model_name=model_name)
# Convert elements to JSON and process the JSON file
elements_to_json(elements, filename=f"{filename}.json")
process_json_file(f"{filename}.json")
# Load the processed text file
...The process_json_file function reads the JSON file generated by elements_to_json, extracts the HTML content of each table element, and writes it to a text file.
The extract_tables_to_docs function uses partition_pdf to extract the tables, converts them to JSON format using elements_to_json, and then calls process_json_file to extract the table HTML. The resulting HTML is saved to a text file.
Converting Tables to Documents
Next, we need to convert the extracted tables into a format that can be ingested by LangChain. We'll split the tables into chunks and create a vector database.
from langchain.text_splitter import CharacterTextSplitter
from langchain.vectorstores import Chroma
from langchain.document_loaders import TextLoader
from langchain.embeddings.openai import OpenAIEmbeddings
def extract_tables_to_docs(filename, output_dir, strategy="hi_res", model_name="yolox", chunk_size=1000, chunk_overlap=200):
...
# Load the processed text file
text_file = f"{output_dir}/{filename.split('/')[-1]}.json.txt"
loader = TextLoader(text_file)
documents = loader.load()
# Split the documents into chunks
text_splitter = CharacterTextSplitter(chunk_size=chunk_size, chunk_overlap=chunk_overlap)
docs = text_splitter.split_documents(documents)
text_file = open(text_file).read()
return docs, text_fileWe use TextLoader to load the processed text file containing the table HTML. We then split the text into chunks using CharacterTextSplitter, specifying the desired chunk size and overlap.
The function returns both the chunked documents (docs) and the raw text content (text_file) for later use.
Displaying Extracted Tables
After extracting the tables from the PDF, we provide an option for the user to view the extracted tables in the Streamlit app.
def main():
...
if uploaded_file is not None:
...
docs, text_file = extract_tables_to_docs(filename, output_dir)
if st.button("Show Tables"):
st.markdown(text_file, unsafe_allow_html=True)
...
In the main function, after the tables are extracted and processed, we create a "Show Tables" button using st.button. When the button is clicked, we use st.markdown to display the raw HTML content of the extracted tables (text_file).
The unsafe_allow_html=True parameter is set to allow rendering of the HTML content. This enables the user to view the extracted tables directly in the Streamlit app.
Asking Questions
Finally, we allow the user to ask questions about the content of the PDF. We use LangChain's RetrievalQA to find relevant chunks from the vector database and generate an answer.
from langchain.chat_models import ChatOpenAI
from langchain.chains import RetrievalQA
os.environ['OPENAI_API_KEY'] = "your_openai_api_key"
embeddings = OpenAIEmbeddings()
llm = ChatOpenAI(model_name="gpt-3.5-turbo", temperature=0)
def main():
...
if uploaded_file is not None:
...
db = Chroma.from_documents(docs, embeddings)
qa_chain = RetrievalQA.from_chain_type(llm, retriever=db.as_retriever())
question = st.text_input("Ask a question about the document:")
if question:
result = qa_chain({"query": question})
st.write("Answer:")
st.write(result["result"])We create a Chroma vector database (db) from the chunked documents (docs) and the OpenAI embeddings. We then initialize a RetrievalQA chain with the OpenAI language model (llm) and the database retriever.
We provide a text input for the user to ask a question about the document. When a question is entered, we pass it to the RetrievalQA chain using qa_chain({"query": question}). The chain searches the vector database, finds relevant chunks, and generates an answer. We display the answer using st.write.
Conclusion
This is how you can get started to build a Streamlit app that extracts tables from PDFs, converts them into searchable documents using vector embeddings, and allows users to ask natural language questions which are answered by an AI language model. You can access the full code by visiting our Github page.
This demonstrates the power of combining Streamlit's simple web app framework with cutting-edge NLP libraries like LangChain and Unstructured. With just a few dozen lines of Python, we built an interactive app that can provide insights from unstructured PDF data.
Some potential enhancements could include handling other file types besides PDFs, allowing users to tweak the AI model's parameters, and providing more visualization options for the extracted data. But even in its basic form, this app showcases an exciting application of AI and data science.
Cohorte Team
December 16, 2024

